RESEARCH HIGHLIGHTS
Our research highlights serve as a collection of feature articles detailing recent scientific achievements on GCS HPC resources.
Researchers Seek A Quantum Leap in Biomedical Research
Research Highlight –
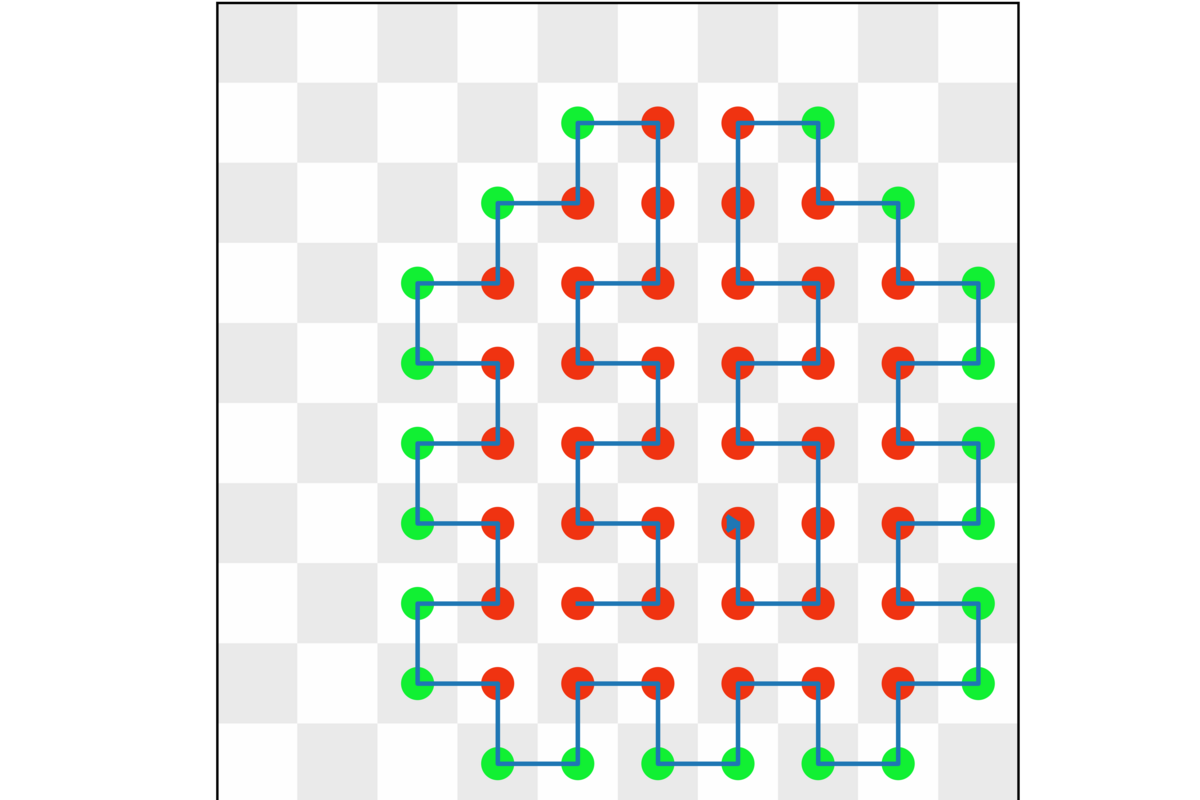
Using JUPSI, the D-Wave Advantage quantum annealer at JSC, the procedure described by Mohanty and his collaborators in Irbäck et al. finds the ground states of chains up to 64 amino acids with a 100% success rate. Here is an example result of a quantum annealing run with 64 amino acids. Red indicates hydrophobic (H) and green represents polar (P) amino acids. Image credit: Sandipan Mohanty
For years, researchers have used the power of high-performance computing (HPC) to advance our understanding of the human body and its interactions with both pathogens and pharmaceuticals. Among other applications, scientists have used simulation to better understand how the protein molecules that perform many important functions in our bodies arrange themselves into specific shapes and how that can influence sickness and health.
While traditional supercomputers are capable of efficiently performing calculations to study how proteins interact with other particles in their environments, studying how protein molecules arrange themselves is still a time-consuming process. Additionally, combing through large databases to find disparate molecular connections between protein and compounds used in medication can still prove difficult, and there are still many humans do not fully understand regarding how proteins’ shapes influence disease or medicine efficacy.
Staff in the Biology Simulation Data Laboratory (SDL) at the Jülich Supercomputing Centre (JSC) are no strangers to working in multidisciplinary groups at the frontier of computational science, often with an eye toward improving medical outcomes. One of the team’s strengths is to push the limits of what is computationally possible. Researchers in the SDL began experimenting with Forschungszentrum Jülich’s (FZJ’s) newest computational tool—the D-Wave Advantage quantum annealer, the largest quantum computer outside of North America—in the hopes that this exciting, novel technology could further accelerate medical research.
“We started learning about what to do and how to translate scientific questions onto this new machine. We realized it is a long journey if you try to do realistic protein simulations, and we really wanted to first understand what the machine can do generally,” said Dr. Sandipan Mohanty, physicist and member of JSC’s Biology SDL. Mohanty and his collaborators at FZJ and Lund University in Sweden developed a reduced model of protein folding as a test case suitable for the current generation of quantum computers. It could also be run on a traditional HPC system to test whether JUPSI could deliver the same results in a shorter timeframe. The team was surprised to learn that in their proof-of-concept study, the quantum computer was not only able to keep up with classical computing; it was able to outperform traditional HPC in certain interesting metrics. The team published its results in Physical Review Research.
Opening the black box
Proteins are made of long flexible chains of amino acids, and the exact sequence of amino acids encodes all information a specific protein needs to fold into a specific three-dimensional shape, which enables the protein to perform its task in the body. When simulating protein folding computationally, researchers must consider astronomically large numbers of different shapes the protein chain might assume, while also considering how the many particles interacting with a protein in its environment may influence its behavior. Ultimately, researchers want to understand how a protein looks in its most probable state for a given set of conditions, such as temperature, pH, and the presence of other molecules in its environment.
Mohanty and his collaborators, who have deployed classical HPC simulations of biomolecules for decades, knew they could not yet mimic all of the complexity contained in such a simulation when using a quantum annealer—the machine is still small compared to world-class classical systems like JSC’s JUWELS supercomputer—but wanted to test its ability to do this work quickly and accurately nonetheless.
To that end, they considered an extremely reduced protein representation, where each amino acid is represented as a single bead along a chain. The beads were only allowed to occupy positions on a 2-dimensional lattice. Amino acids were classified into only 2 types, hydrophobic (H) or polar (P), in contrast to the 20 naturally occurring types of amino acids. The researchers formulated protein folding in this reduced model on the D-Wave quantum annealer. For comparable classical simulations, they used a method based on statistical physics and stochastic importance sampling called “simulated annealing Markov Chain Monte Carlo”. For comparable classical simulations, they used a method based on statistical physics called “simulated annealing Markov Chain Monte Carlo.” With the classical approach, researchers are guaranteed to yield the correct answer when given infinite time to solve the problem, but the shorter the run, the higher the error rate.
A powerful laptop or small cluster can perform a modest Monte Carlo simulation, such as this model protein experiment. But the longer the chain of amino acids, the more computationally expensive it becomes to keep the estimated errors small, meaning researchers must have access to increasingly long periods of computational time or settle for less accuracy. When doing a chain of 30 amino acids, for instance, classical simulated annealing can reach 80 percent accuracy in finding the ground state taking about as much time as a quantum annealer needs for an annealing cycle, except that the quantum annealer seeks out the ground state with 100 percent accuracy. When simulating a chain of 48 or 64 amino acids, the researchers found that when running for a similar amount of time, the classical machine would only deliver 10–15 percent accuracy while the quantum machine remained at 100 percent.
“When you are doing Monte Carlo simulations, you are sampling more and more in order to get a finer and finer approximation of the results. Getting to a 100 percent success rate in the search for the ground state using classical simulated annealing requires running the annealing cycles slowly over longer periods of time,” Mohanty said. “The biggest surprise for us in this study was that the quantum annealer can get to that accurate approximation using far smaller resources. The classical simulation incurs the computational cost of finding and evaluating a long series of states one after the other. In contrast, the quantum annealing process behaves as if it examines all possible states at the same time and zeros in on the ground state.”
Next-generation computing needs world-class HPC centers
Mohanty reiterated that despite the team’s significant accomplishment, the simulation is still a far cry from what is currently possible on traditional HPC resources. In fact, he reiterated that now and in the future, traditional HPC resources will remain an essential tool and that quantum computing needs to be accurately placed in the overall landscape of computing technologies available for research.
“When we are exploring or coming face-to-face with a new technology, we have a tendency to downplay the achievements of the previous generation of technology,” he said. “I’ve seen some of that when talking about quantum computing, and that has the danger of overpromising. When people don’t have their expectations met—whether they were realistic or not—they see a new technology as a bust. I think classical HPC systems and quantum systems are both going to have an important role to play in research moving forward.”
Mohanty cautioned that it was too early to claim what exact role quantum computers would carve out in the increasingly diverse hardware landscape at HPC centers but indicated that centers like JSC served an indispensable role in advancing the state-of-the-art in computational sciences. He indicated that the Gauss Centre for Supercomputing (GCS) centers’ staffs all have different forms of expertise, and the close exchange between experts helps drive innovation.
“Our role is not looking at computational science from a high level, where everything is just a black box that we take and use,” he said. “We, as researchers in a scientific domain, do sometimes use large computing systems in the same way that many people drive a car—you don’t always have to open the hood to understand how it all works in order to go where you want to go. However, at our institute, we try to push new technologies, sometimes even beyond what it can actually do, so we can learn how a machine does something and come away with new and interesting insights.”
-Eric Gedenk
Related Publication: Irbäck, A., L. Knuthson, S. Mohanty, and C. Peterson. “Folding Lattice Proteins with Quantum Annealing,” Physical Review Research 4 (2023). DOI: 10.1103/PhysRevResearch.4.043013