LIFE SCIENCES
Continuous, stable processes for the sustainable enzymatic production of chiral amino alcohols integrating downstream processing
Principal Investigator:
Prof. Dr. Holger Gohlke
Affiliation:
Heinrich-Heine-Universität Düsseldorf, Institut für Pharmazeutische und Medizinische Chemie, Germany
Local Project ID:
metapro
HPC Platform used:
JUWELS BOOSTER at JSC
Date published:
Teaser
Prof. Dr. Holger Gohlke and his team used the compute resources of the Jülich Supercomputing Centre to study the carboligase ApPDC and the transaminase Cv2025, enzymes of high industrial interest, as biocatalysts to produce fine chemicals to be used as agrochemicals or pharmaceutical compounds. The team performed extensive molecular dynamics simulations for enzyme variants in different solvents compositions and used Constraint Network Analysis (CNA) to simulate the thermal unfolding process of the enzymes in a rigid cluster decomposition. The computational results revealed significant differences in protein stability and unfolding, and in small molecules repartitions and binding. In the context of ApPDC, computational predictions were the key for the suggestion of novel thermostable and process-stable variants. Together, these findings showcase the importance of combining computational with experimental models to address enzyme stability bottlenecks, paving the way for identifying further viable bioprocesses in green chemistry.
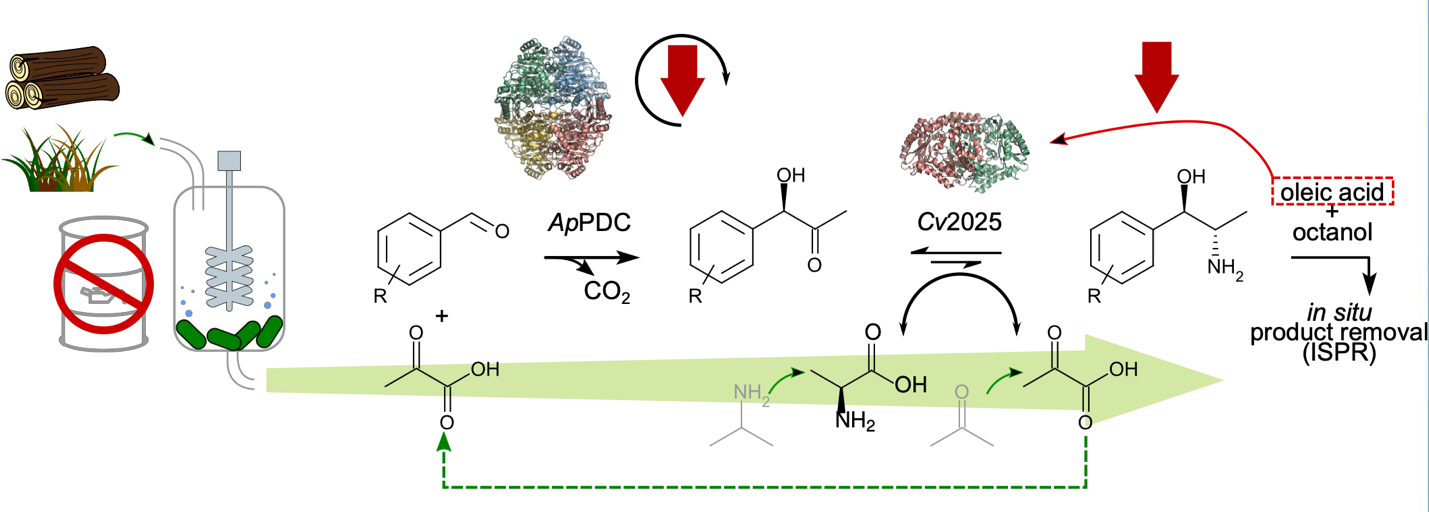
Figure1. Metaraminol production achieved by an integrated process using second-generation feedstocks, in a two-step reaction performed by a carboligation by ApPDC and a transamination by Cv2025. Both biocatalysts have been shown to be inactivated throughout the process. Here, reactive extraction is mandatory to access high product concentrations of the chiral amino alcohol. The optimal operation mode of the transformation starting from 3-OH-benzaldehyde and pyruvate is expected to be a cascaded continuous mode or repetitive fed-batch.
Project
Chiral amino alcohols are critical intermediates in fine chemical and pharmaceutical synthesis. The enzymatic production of these compounds via cascaded biocatalysts can improve sustainability but requires enhanced enzyme stability and activity under process conditions. The BioSC MetaProcess project targets this challenge through combined computational and experimental methods to develop stable enzyme variants and optimize processing environments.
In this project, Prof. Dr. Gohlke and Rocco Gentile, Dr. Jesko Kaiser, Dr. Daniel Becker, and Dr. Stephan Schott-Verdugo focused on the Acetobacter pasteurianus pyruvate decarboxylase (ApPDC), catalyzing the carboligation step, and the Chromobacterium violaceum transaminase Cv2025, responsible for the subsequent transamination (Figure 1). Both enzymes show limitations: ApPDC suffers inactivation during reaction cycles, and Cv2025 activity is inhibited by oleic acid used in the reactive extraction process to obtain the product.
To generate ApPDC stable enzyme variants, AI-assisted tools, such as ProteinMPNN, SaProt, and ESMwere used to guide mutational design. Among predicted variants, HG09, HG10, HG11, and 4mut_HG09 were selected for improved process stability based on consensus stability predictions and folding free energy assessments with molecular simulations and CNA analysis. The 4mut_HG09 is the combination of the thermostable variant 4mut (which derives from the published PDC-Var.2) and the HG09 variant. These variants demonstrated prolonged activity retention during extended incubations and improved thermal stability (for the combined 4mut_HG09), indicating enhanced chemical stability and possible allosteric effects. Molecular dynamics simulations and rigidity analyses confirmed differential stabilization patterns across monomers, guiding future engineering strategies.
For Cv2025, free ligand diffusion simulations mapped oleic acid binding sites, used in in situ product removal, correlating with potential enzyme inhibition patterns. These computational insights link molecular interaction mechanisms to process challenges, informing enzyme redesign or process adjustments to mitigate deactivation.
As a potential way to address stability issues of enzymes, molecular simulations were performed and contrasted with experiments in a micro-aqueous reaction system (MARS), where cyclopentyl methyl ether (CPME) with minimal amounts of water are used as reaction condition. Solvent-enzyme interactions affecting the activity and stability of ApPDC were identified, revealing substrate and product repartitioning that could lead to improved catalytic conditions.
The team generated structural models for the predicted variants and solvent conditions using FoldX and performed Constraint Network Analysis on large ensembles extracted from extensive molecular dynamics simulations. Solvated simulation boxes for all systems were created using the AmberTools22 software package, leading to system sizes of 45’000 atoms. The team used the GPU particle mesh Ewald implementation of the AMBER22 molecular dynamics simulation suite to perform a combined total of ~500 μs of simulation time over the simulation sets, which was made possible using the highly efficient A100 GPUs on the JUWELS Booster partition.
Overall, this integrated approach combining enzyme stabilization through variant design and solvent engineering provides a scalable strategy for sustainable production of chiral amino alcohols, advancing continuous cascade biocatalysis platforms.