LIFE SCIENCES
Fitting Multi-Cellular Stochastic Models Using Approximate Bayesian Computation
Principal Investigator:
Prof. Dr. Hasenauer
Affiliation:
Universität Bonn, Hausdorff Center for Mathematics - HCM, Bonn, Germany
Local Project ID:
fitmulticell
HPC Platform used:
JUWELS of JSC
Date published:
Abstract
FitMultiCell is a computational pipeline developed by Prof. Dr.-Ing. Jan Hasenauer's team to tackle the complexity of simulating and fine-tuning biological tissues. This tool streamlines the creation, simulation, and calibration of biological models that imitate cellular interactions within tissues. The pipeline offers a user-friendly platform for researchers to conduct analyses on supercomputers like JUWELS. FitMultiCell's flexibility and power are demonstrated in studies on viral infections, tumor growth, and organ regeneration, proving its efficiency in refining models to match experimental data. Furthermore, enhancements for handling data outliers and scalability ensure FitMultiCell's robust application in diverse research fields.
Report
In the process of understanding the complex mechanisms that govern biological tissues, scientists usually use computational models. These models try to simulate the interaction of cells with their environments, which is an essential step to understanding how these tissues develop, maintain themselves, and respond to various challenges, including diseases.
These tissues are always active, where cells can interact in many different ways, like sending signals, regulating genes, and touching each other. These interactions are complex, happening on different scales, both small (molecular) and large (cellular). To model these processes on a computer, we need to understand the rules that guide how cells behave. Then, we use this rule to build the computational model. However, one of the main challenges to building a representative model is called model calibration: the process of adjusting the settings or parameters of a model so that its predictions become an accurate representation of reality (similar to tuning musical instrument, to ensure it plays the right notes). A team led by Prof. Dr.-Ing. Jan Hasenauer introduced a pipeline called FitMultiCell [1], an innovative computational framework designed to address this challenge by facilitating the simulation and calibration of multi-scale and multi-cellular models.
FitMultiCell is a tool that helps solve the above issue. It's a free framework that brings together model construction, simulation of the model, and model tuning together into one smooth process. It uses two advanced tools: Morpheus for constructing the model and creating simulations, and pyABC for parameter fine-tuning. In addition to that, FitMuliCell allows scientists to use large computer systems, allowing for the study of many complex biological activities.
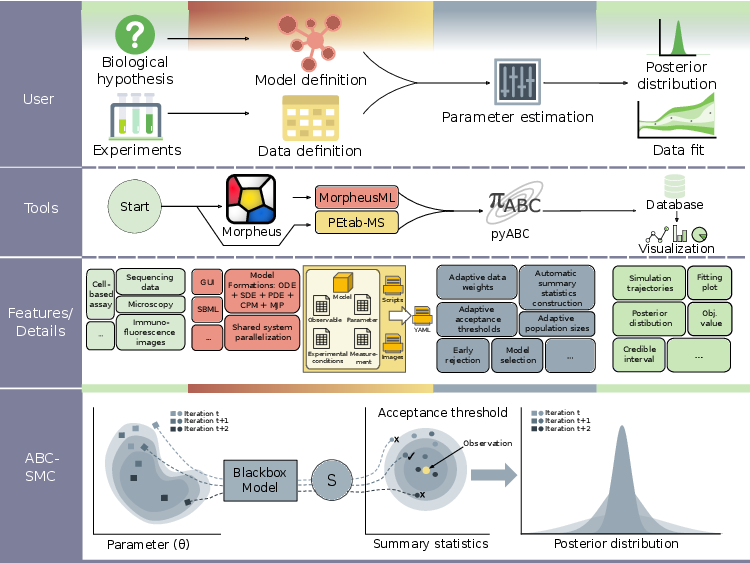
The user-friendly nature of FitMultiCell is one of its key strengths. It provides a modular framework that simplifies the entire process, from model formulation to parameter estimation and analysis. Researchers can interact with the pipeline through a graphical user interface (GUI) or by using Python coding, offering flexibility to suit different user preferences and programming expertise levels. This accessibility is important for increasing the usability of the pipeline and enabling a wide range of users' spectrum to investigate the multi-cellular processes without the need for deep computational expertise.
The applicability of FitMultiCell is tested and demonstrated in multiple application examples and case studies, including the simulation of viral infections, tumor growth, and organ regeneration. All these applications were implemented, tested, and simulated at the JUWELS cluster at the Jülich Supercomputing Centre (JSC). Such a powerful system is needed since millions of simulations are being conducted to ensure better model calibration of these complex models. Additionally, the ability to run simulations on high-performance clusters like JUWELS, with its large computational capability, allows for scalability and faster results, making FitMultiCell a valuable tool for researchers in various fields.
These examples show that not only the pipeline is capable of accurately simulating complex biological processes but also its effectiveness in adjusting model parameters to align with experimental observation and data, all in an efficient manner. By doing so, FitMultiCell proves to be a valuable tool for validating biological hypotheses, exploring the effects of different perturbations, and understanding the underlying mechanisms of disease progression and treatment responses.
As these computational demands remained high, the team of Prof. Hasenauer also looked into the possibility of improving the scalability of such a pipeline further. Their effort was rewarded with the development of a new scalable [2] scheme that has proven to be both accurate and efficient when compared to existing schemes.
Their effort was also extended to developing an automated system to manage how the pipeline was orchestrating itself in these large clusters of compute nodes [3]. This system organizes work into three main parts: a central server that assigns tasks, multiple worker processes ready to carry out the simulation tasks, and a parameter-tuning task that runs the actual inference analysis. This setup ensures that tasks are distributed evenly and efficiently, to ensure optimal performance.
The team was interested in improving the performance of FitMultiCell, but also its flexibility, especially when dealing with complex data like images. Application problems with imaging data pose a particular challenge as it is crucial to extract the essential information (but not more) to achieve simultaneously a high accuracy and performance. The team investigated the use of some well-known methods that simplify data to its most important aspect but found some issues. To overcome these issues, they improved the methods by adjusting how much attention was given to different parts of the data, focusing on the most rewarding information, and using a detailed analysis to get clearer results. They found a combination of these improvements worked best for making sense of complex data. All the tests conducted to reach to the conclusion were carried out on the same JUWELS supercomputer [4].
To improve the pipeline further, the team led by Prof. Hasenauer investigates data outliers, or unusual data points, which are data that are significantly different from the rest of the data and can skew results if not properly addressed. By identifying and removing outliers, researchers can ensure their analysis is more accurate and reliable. However, their presence can mess up statistical analysis, which is an important step in model calibration. To address this, the team developed unique functions that are robust to outliers. These functions proved to be particularly useful for complex data like images. All tests on the JUWELS supercomputer showed that the new methods are much better at handling tricky data [5].
In conclusion, FitMultiCell represents key progress in the field of computational biology. By providing a scalable and user-friendly pipeline for the simulation and calibration of multi-cellular models, it enables researchers to develop a better understanding of biological tissues. This will allow for a better understanding of cell behaviour and tissue dynamics. As computational modelling becomes increasingly essential to biological research, tools like FitMultiCell will make researchers focus on testing hypotheses and interpreting results, rather than spending time on technical challenges.
References
- Emad Alamoudi, Yannik Schälte, Robert Müller, Jörn Starruß, Nils Bundgaard, Frederik Graw, Lutz Brusch, Jan Hasenauer, FitMultiCell: simulating and parameterizing computational models of multi-scale and multi-cellular processes, Bioinformatics, Volume 39, Issue 11, November 2023, btad674, https://doi.org/10.1093/bioinformatics/btad674
- Emad Alamoudi, Felipe Reck, Nils Bundgaard, Frederik Graw, Lutz Brusch, Jan Hasenauer, and Yannik Schälte. A Wall-time Minimizing Parallelization Strategy for Approximate Bayesian Computation." arXiv preprint arXiv:2305.00506 (2023).
- Emad Alamoudi, Jörn Starruß, Nils Bundgaard, Robert Müller, Felipe Reck, Frederik Graw, Lutz Brusch, Jan Hasenauer, and Yannik Schälte. Massively Parallel Likelihood-Free Parameter Inference for Biological Multi-Scale Systems. volume 51 of Publication Series of the John von Neumann Institute for Computing (NIC), pages 355 – 365, Julich, 2022. Forschungszentrum Jülich GmbH Zentralbibliothek, Verlag
- Yannik Schälte, and Jan Hasenauer. Informative and adaptive distances and summary statistics in sequential approximate Bayesian computation. Plos one 18.5 (2023): e0285836.
- Yannik Schälte, Emad Alamoudi, and Jan Hasenauer. Robust adaptive distance functions for approximate Bayesian inference on outlier-corrupted data. bioRxiv (2021): 2021-07.