LIFE SCIENCES
MD Simulations of Large Membrane Systems: From Membrane Protein Arrays to the Influenza Virus
Principal Investigator:
Mark S.P. Sansom
Affiliation:
University of Oxford (Great Britain)
Local Project ID:
PP12061115
HPC Platform used:
Hermit of HLRS
Date published:
Membrane proteins are of great biomedical importance. They account for ~25% of all genes and are involved in diseases ranging from diabetes to cancer. Membrane proteins play a key role in the biology of infection by pathogens, including both bacteria and viruses. They also play an important role in signalling within and between cells. It is therefore not surprising that membrane proteins are major targets for a wide range of drugs and other therapeutic agents. Recently, the number of known structures of membrane proteins has started to increase. Large scale computer simulations allow researchers to study the movements of these proteins in their native membrane environments.
However, biologically realistic simulations of the dynamic behaviour of membrane proteins require calculations on millions of atoms and therefore the use of advanced supercomputers are essential for studying their complex dynamic behaviour.
A highly realistic computational model of the human influenza virus was constructed in the research group. The project which was made possible through the European HPC initiative PRACE (Partnership for Advanced Computing in Europe) enabled the scientists to simulate this flu virus model in water droplets at different temperatures. As flu is a seasonal virus in temperate climates, this data provides the researchers with molecular-level insight into the biophysical properties of the virus under conditions that are of substantial impact to society. It remains unclear why the flu virus exhibits a seasonal infectivity fluctuation and our the results achieved provide a uniquely high-resolution insight into the interaction of proteins and lipids at different temperatures in this system. Thus the researchers were granted computing time on GCS petascale system Hermit of HLRS to simulate detailed aspects of a full-scale life form/pathogen. These massive (~5.6 million particle) simulations would be simply impossible without advanced computational facilities such as provided by GCS centre HLRS Stuttgart.

Copyright: © University of Oxford, Dept. of Biochemistry
The scientists also conducted extensive simulations of models of crowded membranes of the bacterium E. coli, and of mammalian cells. Both through the E. coli membrane models and more complex mammalian membrane they explored the curving and bending fluctuations occurring within such membranes. Because of the data obtained as a result of these simulations, the scientists can now start to understand how local fluctuations in the shape of cell membranes may influence biological functions.
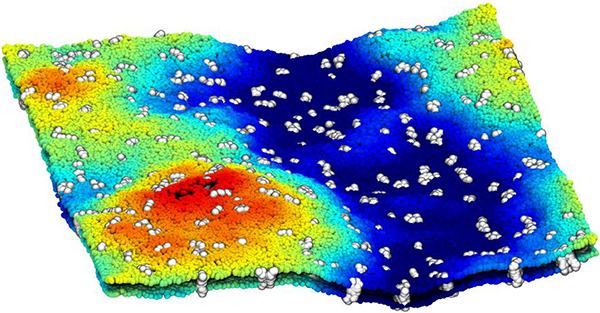
Copyright: © University of Oxford, Dept. of Biochemistry
The scientific team undertaking this research reflected the international and multidisciplinary nature of research, and consisted of Prof. Mark Sansom (team leader), Dr. Joseph Goose, Dr. Tyler Reddy, Dr. Heidi Koldsø, Dr. Philip Fowler, Dr. Antreas Kalli, and Mr. Jean Hélie.
Scientific Contact:
Prof. Mark S.P. Sansom
David Phillips Professor of Molecular Biophysics & Head of Department,
Dept. of Biochemistry, University of Oxford, South Parks Road, Oxford, OX1 3QU/Great Britain
e-mail: mark.sansom@bioch.ox.ac.uk