LIFE SCIENCES
Structural Determinants in the Activation/Permeation of Plant Glutamate Receptor-Like (GLRs) Receptors
Principal Investigator:
Prof. Dr. Holger Gohlke
Affiliation:
Heinrich-Heine-Universität Düsseldorf, Institut für Pharmazeutische und Medizinische Chemie, Düsseldorf, Germany
Local Project ID:
glr
HPC Platform used:
UWELS BOOSTER at JSC
Date published:
Teaser
Plants may not have brains, but they do have glutamate receptor–like proteins (GLRs) that behave much like the ion channels driving learning and memory in animals. These mysterious channels regulate key plant functions, from nitrogen use to pollen growth, yet how they control ion flow has remained unclear. To uncover their secrets, researchers led by Prof. Dr. Holger Gohlke combined high-precision modeling and molecular dynamics simulations of moss GLRs. Using AlphaFold2, they built accurate 3D channel structures and revealed how subtle changes in pore residues reshape ion permeability. The mutant channel showed enhanced calcium flow, linked to a more electronegative pore, an insight confirmed by large-scale simulations on supercomputers. These findings shed light on how plant GLRs balance cation and anion transport, bringing scientists closer to understanding how plants use animal-like ion channels to coordinate their complex physiology.
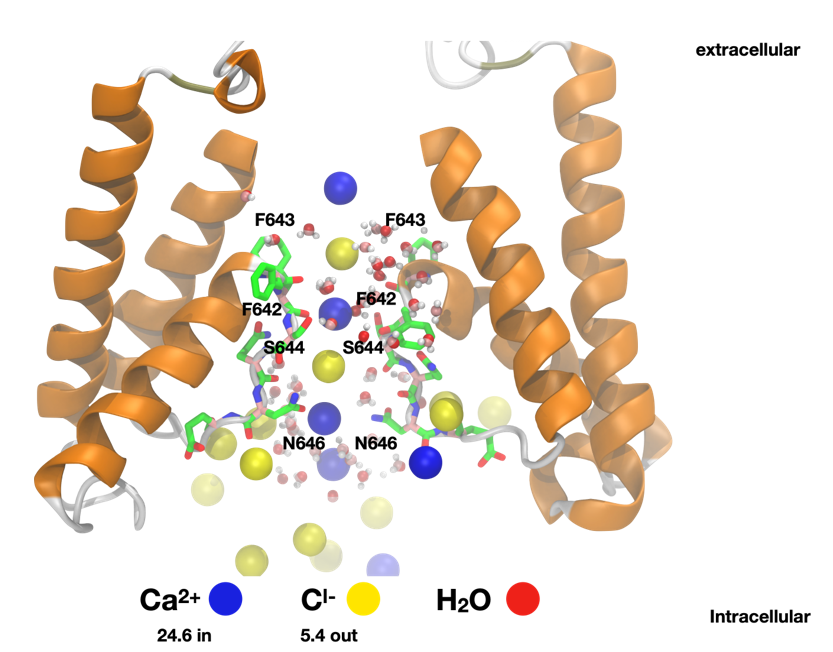
Figure 1. Ion permeation mechanism in GLRs. Residues involved in ion coordination at the selectivity filter (SF) are shown in a licorice representation. Ca2+ (blue) and Cl- (red) ions are shown as spheres. The SF is shown in a cartoon representation.
Project
Ionotropic glutamate receptors (iGluRs) are ion channels that open in response to the neurotransmitter glutamate, playing essential roles in learning and memory in animals. Surprisingly, plants also possess glutamate receptor–like proteins (GLRs), even though they lack a nervous system. These plant GLRs are involved in diverse physiological processes such as nitrogen metabolism, gas exchange, immune responses, and pollen tube growth. However, their molecular mechanisms, especially how they are gated by ions, which ligands activate them, and how they select specific ions, remain poorly understood. To bridge this gap, researchers are combining experimental and computational approaches to model and simulate GLR structure and function. Recent findings from moss GLR studies suggest that plant GLRs exhibit unusually broad ion selectivity, and engineering their pore regions using animal receptor motifs can enhance ion permeability and gating, offering new insights into how these unique plant ion channels operate.
Understanding the permeability of plant glutamate-like receptors (GLRs) is key to uncovering their physiological roles. Unlike animal ionotropic glutamate receptors (iGluRs), which are selective for positively charged ions, plant GLRs can transport both cations and anions. To explore the structural basis of this dual permeability, the team around Prof. Dr. Holger Gohlke performed extensive molecular dynamics (MD) simulations under applied electric fields to model ion flow through the channels. Initially, building on earlier work, the team reconstructed 3D models of the moss channel PpGLR1 and a mutant version, PpGLR1-SWAP, using the full AlphaFold2 platform. These models, focused on the transmembrane domain (TMD), underwent rigorous refinement to ensure high structural quality, forming the basis for subsequent simulations. Structural validation against the experimentally resolved Arabidopsis GLR3.4 confirmed that AlphaFold2 produced the most accurate TMD representations. While earlier ColabFold models showed inconsistencies in membrane helix orientation, the AlphaFold2 models correctly aligned all transmembrane segments (M1–M4), allowing for reliable structural and electrostatic analyses.
Electrostatic potential mapping then revealed that the PpGLR1-SWAP variant possessed a more electronegative pore region than the wildtype, particularly near residues Q643 and Q644—positions known to control cation selectivity in animal receptors. This finding suggests a molecular explanation for the increased cation permeability observed experimentally in the mutant channel. All-atom MD simulations performed on the Juwels Booster GPUs confirmed that both channels permit the passage of calcium and chloride ions, consistent with experimental data. Detailed analyses identified key residues, F642, F643, S644, N646, and D647, as central to ion stabilization and transport. The mutant channel displayed stronger calcium binding near the selectivity filter, while the wildtype favored chloride permeation.
Overall, these achievements mark major progress toward understanding ion selectivity in plant GLRs. The ongoing refinement of structural models and simulations promises to deepen insights into how these unique plant ion channels function at the molecular level.