LIFE SCIENCES
The Allosteric Effect of the SH2 Domain on Abl Kinase Activation
Principal Investigator:
Francesco Luigi Gervasio
Affiliation:
Department of Chemistry and Institute of Structural Molecular Biology, University College London (UK)
Local Project ID:
pr86ga
HPC Platform used:
SuperMUC of LRZ
Date published:
Protein kinases are the key enzymes that control most cellular activities. A kinase that fails to work properly can therefore cause severe damage to the organism, causing diverse diseases including cancer. It is therefore highly desirable to develop drugs that modulate the activity of specific protein kinases. However, this task is complicated by the fact that all protein kinases share a common architecture, so a drug tailored to one kinase is likely to affect others as well, causing serious side effects.
The Abelson tyrosine kinase (Abl) is of special interest because of its importance as an anti-cancer drug target. A genetic defect affecting the gene that encodes Abl causes chronic myeloid leukemia (CML), and Abl inhibitors are the only known anti-CML drugs. The treatment is very effective, but a significant proportion of patients relapses due to drug resistance-causing mutations in Abl.
Activation of Abl involves a complete rearrangement of the domains of the protein, and thus the interface between the catalytic and the modulator (SH2) domains seems to be a promising target for new anti-CML drugs. In a project, which was made possible through the Partnership for Advanced Computing in Europe (PRACE) using the supercomputing infrastructure of the Leibniz Supercomputing Centre in Garching near Munich, a team of scientists set out to shed light on the effect of the SH2 domain on the intrinsic motions of the catalytic domain.
The researchers used Molecular Dynamics simulations, in which all atoms of the system are represented by rigid spheres, and interactions between them are described by simplified models. Integration of Newton's equations of motion then yields a trajectory describing the movements of the system. The integration is carried out in tiny time steps in the range of femtoseconds, which have to be repeated millions of times. The simulation of meaningful protein motions therefore requires massive computer power.
The simulations indicate that the SH2 domain modifies the motions of the catalytic domain of Abl in two ways. On the one hand, it stabilizes and positions some elements of the enzyme in an optimal arrangement, and on the other hand it channels thermal fluctuations into global movements involved in the catalytic process.
In order to validate this model, in collaboration with the group of Prof. Giulio Superti-Furga at CeMM in Vienna, the scientists designed a set of mutants and tested them experimentally for their catalytic activity. It is expected that in the future these findings may open the door to the development of anti-CML drugs with a new mechanism of action and therefore effective for the treatment of patients who have become resistant to the current treatments.
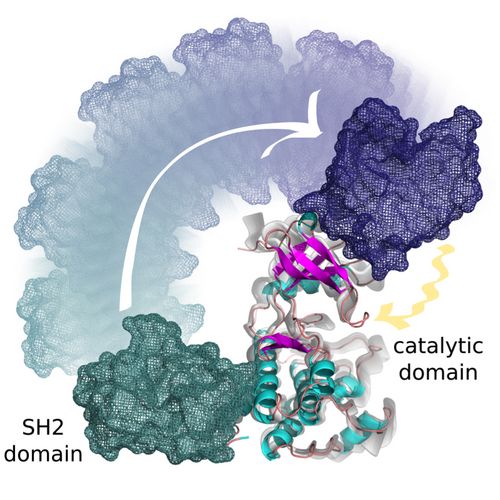
Figure 1: Activation of the Abl kinase by its SH2 domain. The SH2 domain migrates from the lower C-terminal to the upper N-terminal part of the catalytic domain, where it modifies the structural arrangement of protein elements involved in catalysis and controls the global motions of the catalytic domain. © Spanish National Cancer Research Center (CNIO), Madrid, Spain
The results of this project were recently published in:
N. Dölker, M. Gorna, L. Sutto, A. Sanchez Torralba, G. Superti-Furga, and F. L. Gervasio. The SH2 domain regulates c-Abl kinase activation by a cyclin-like mechanism and remodulation of the hinge motion. PLOS Comp Biol, 10(10):e1003863, 2014.
Scientific Team:
Francesco L. Gervasio (Principal Investigator)1,2, Nicole Dölker1, Ludovico Sutto1,2, Giorgio Saladino1,2, Silvia Lovera1,2
1) Spanish National Cancer Research Center (CNIO), Madrid, Spain.
2) Now at: University College London, UK
Scientific Contacts:
Prof. Francesco Luigi Gervasio, PhD
Chair in Bio-molecular Modeling
Department of Chemistry and Institute of Structural Molecular Biology
University College London
20 Gordon Street
London WC1H 0AJ, UK
e-mail: f.l.gervasio @ ucl.ac.uk
Dr. Nicole Dölker
Centro Nacional de Investigaciones Oncológicas
Structural Computational Biology Group
C/ Melchor Fernández Almagro, 3
28029 Madrid/Spain
e-mail: andolker @ cnio.es
LRZ project ID: pr86ga
January 2015