LIFE SCIENCES
Towards the Construction of a Large Foundational Model for Protein Structures with Message-Passing Graph Neural Networks
Principal Investigator:
Prof. Dr. Holger Gohlke
Affiliation:
Heinrich-Heine-Universität Düsseldorf, Institut für Pharmazeutische und Medizinische Chemie, Düsseldorf, Germany
Local Project ID:
found
HPC Platform used:
JUWELS BOOSTER module at JSC
Date published:
Teaser
Deep learning is revolutionizing protein science, with graph neural networks (GNNs) and multimodal models enabling unprecedented insights into protein function and design. In this project, the team led by Prof. Dr. Holger Gohlke developed two complementary AI models: TopEC and OneProt. TopEC uses 3D GNNs to predict enzyme functions directly from protein structures, incorporating atomic distances and angles to achieve high accuracy across more than 800 enzyme classes. Its structure-aware approach outperforms traditional 2D methods and remains robust even when binding site information is uncertain. In parallel, OneProt extends the multimodal ImageBind framework to proteins, aligning structural, sequence, text, and binding data into a shared representation space. This lightweight fine-tuning strategy enhances retrieval and prediction tasks and reveals evolutionary relationships between proteins. Together, TopEC and OneProt showcase how next-generation AI can accelerate enzyme discovery, drug design, and protein engineering.
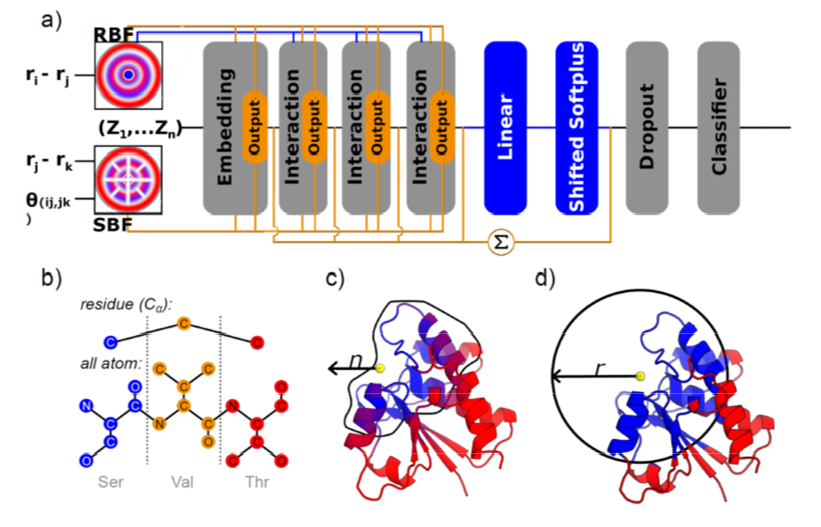
Figure 1. Protein input and neural network architecture of TopEC. a) Overview of the SchNet (blue) and DimeNet++ (orange) architectures, with shared components shown in grey. Both models embed atomic numbers and use filters to encode atomic distances and angles: radial Bessel filters (RBFs) for pairwise distances and spherical Fourier Bessel (SBF) filters for triplet distances and angles. DimeNet++ differs by summing over all embedding and interaction blocks to generate predictions. b) Localized 3D descriptors at different resolutions: in the residue view, only Cα atoms are retained and nodes represent amino acid types; in the all-atom view, each heavy atom forms a node coded by its chemical environment. Blue residues are included in the model input, red ones are excluded. c) Count representation: residues are selected by the n nearest neighbors to a central site. d) Distance representation: residues are selected within a radius r around the binding site center.
Project
Recent advances in deep learning, particularly graph neural networks (GNNs), are transforming the way we analyze and predict protein functions. GNNs can represent the complex 3D architecture of proteins as graphs, capturing atomic interactions and spatial relationships that underlie biological activity. While building such detailed structural graphs is computationally demanding, innovative message-passing networks now enable efficient encoding of distances and angles for larger biomolecules. At the same time, the integration of multiple data modalities, such as sequence, structure, and binding information, is emerging as a powerful strategy to generate richer protein representations. Combining these multimodal approaches with advanced neural architectures promises to enhance predictions of protein function, stability, and molecular interactions—paving the way for more accurate enzyme and antibody design.
Accordingly, in this project, the team around Prof. Dr. Holger Gohlke pursued the development of TopEC, which uses Graph Neural Networks for enzyme function prediction, and OneProt, which effectively extends the ImageBind framework to the protein space.
TopEC is a 3D graph neural network based on a localized 3D descriptor to learn chemical reactions of enzymes from enzyme structures and predict Enzyme Commission (EC) classes. Using message-passing frameworks, the team included distance and angle information to significantly improve the predictive performance for EC classification (F-score: 0.72) compared to regular 2D graph neural networks. It trained networks without fold bias that can classify enzyme structures for a vast functional space (>800 ECs). The model is robust to uncertainties in binding site locations and similar functions in distinct binding sites. The team observed that TopEC networks learn from an interplay between biochemical features and local shape-dependent features.
OneProt is a multi-modal AI for proteins that integrates structural, sequence, text, and binding site data. Using the ImageBind framework, OneProt aligns the latent spaces of protein modality encoders in a lightweight fine-tuning scheme that focuses on pairwise alignment with sequence data rather than requiring full matches. This novel approach demonstrates strong performance in retrieval tasks and showcases the efficacy of multi-modal systems in Protein Machine Learning through various downstream baselines, including enzyme function prediction and binding site analysis. Furthermore, OneProt enables the transfer of representational information from specialized encoders to the sequence encoder, enhancing capabilities for distinguishing evolutionarily related and unrelated sequences. In particular, the fine-tuning scheme exhibits representational properties where evolutionarily related proteins align in similar directions within the latent space. This work expands the horizons of multi-modal protein models, paving the way for transformative applications in drug discovery, biocatalytic reaction planning, and protein engineering.
References
van der Weg, K., Merdivan, E., Piraud, M., Gohlke, H.
TopEC: prediction of Enzyme Commission classes by 3D graph neural networks and localized 3D protein descriptor.
Nature Commun. 2025, 16, 2737.
Flöge, K., Udayakumar, S., Sommer, J., Piraud, M., Kesselheim, S., Fortuin, V., Günneman, S., van der Weg, K.J., Gohlke, H., Merdivan, E., Bazarova, A.
OneProt: towards multi-modal protein foundation models via latent space alignment of sequence, structure, binding sites and text encoders.
arXiv: 10.48550/arXiv.2411.04863, 2024.