RESEARCH HIGHLIGHTS
Our research highlights serve as a collection of feature articles detailing recent scientific achievements on GCS HPC resources.
Researchers Use Supercomputers in an Effort to Develop Safer, More Personalized Medical Procedures for Respiratory Illnesses
Research Highlight –
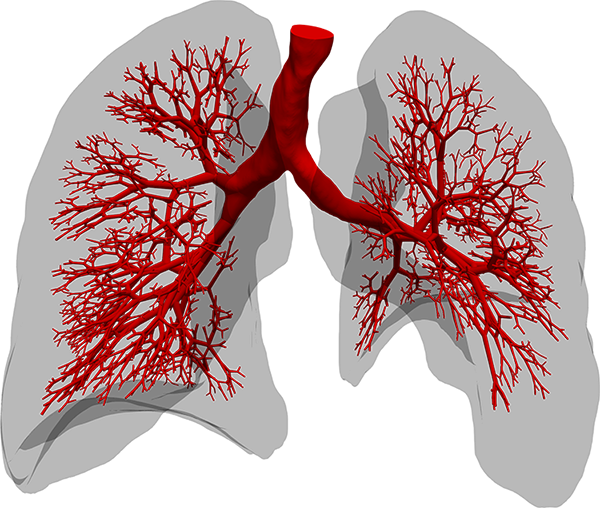
The section of the lung geometry (red) is fully resolved by three-dimensional simulations. From the smallest features in the illustration, the spongy network of the lung continues into even smaller structures where the flow is laminar. The researchers’ current simulation practice applies a reduced model representing the effective mechanical response in terms of a boundary condition specific to each individual airway section. Image Credit: Maximilian Bergbauer
Since March, 2020, the world has confronted a cold reality long known to epidemiologists and other public health experts—viruses that cause novel respiratory illnesses are among our greatest microbial foes. Viruses that attack human lungs can spread easily and leave lasting damage in their wake. The novel SARS-CoV-2 virus, for instance, unleashed the COVID-19 pandemic and has infected hundreds of millions of people, leaving more than 5 million dead.
Part of what makes viruses such as SARS-CoV-2 so dangerous (and so terrifying) is the organ they attack. Zoom in to view human lungs at the micro level, and you see a delicate, sponge-like organ with very thin, flexible and sensitive structures that allow a direct connection to the human circulatory system. The novel virus attacks these vulnerable parts and heavily increases the number of patients suffering from Acute Respiratory Distress Syndrome (ARDS) – a disease that so far has been caused by sepsis, pneumonia, traumata, smoke or toxic gas inhalation, among other causes, and strongly lowers the chances for survival of a person.
Modern approaches for supporting patients fighting off acute respiratory illness, such as inserting a tube into a person’s trachea and using a machine to mechanically assist with breathing, have helped stave off death for COVID patients as well as others with lung damage or injury, but these methods also come with their own risks. In essence, ventilators add stress to an already damaged organ, and prolonged use can lead to complications, other health problems, and frequently even lead to death themselves.
Over the last 15 years, a team of researchers from the Technical University of Munich (TUM) has worked on novel approaches for developing strategies to better assess risks associated with machine ventilation and other invasive health procedures to combat respiratory illness. In the last five years this was supported via collaboration with staff at the Leibniz Supercomputing Centre (LRZ) in Garching near Munich. With the advent of the COVID-19 pandemic, the team’s work became even more essential.
“For the research in our group, COVID-19 didn’t change our focus, per se, because we were working on this issue already,” said Prof. Dr. Wolfgang Wall, Professor of Computational Mechanics and Director of the Institute for Computational Mechanics at TUM as well as the principal investigator on this project. “Of course, the pandemic became something that raised awareness of this issue, and these acute lung injuries are one of the major complications that have led to people dying when having COVID, but the mortality rate for patients with ARDS requiring respirators was already high. In that sense, COVID became a bit of added motivation and visibility for us.”
Using the SuperMUC-NG supercomputer at LRZ and closely collaborating with the centre’s computational experts, the team mixed relatively new computational approaches with modified classical computational fluid dynamics (CFD) techniques, achieving a significant performance boost when doing high-resolution lung modelling. In the coming years, the team will take this approach to design both more accurate as well as novel reduced-order models that could be used by medical professionals to inform how to best mechanically respirate patients. The team presented its research at the SC21 computing conference this November.
Smorgasbord of simulation techniques
Researchers across multiple scientific and engineering disciplines use CFD simulations to help solve difficult problems. From modelling air flow around a wind turbine to gaining increased insight into the physics and chemistry happening in a fuel injector during combustion, CFD allows researchers to gain insights into things either too diffuse or difficult to see experimentally.
The complications in a CFD simulation primarily stem from scales—both size and time. Researchers have to simulate features of a fluid flow at fine enough detail to capture realistic behaviour while simultaneously creating a large enough simulation to reflect how the system would behave in real-world conditions.
The first step typically requires that researchers break up their simulations into a computational grid of small mesh cells. Researchers then solve equations for these individual grid spaces—a matrix, of sorts—that represent how “fluid particles” behave with one another. In order to capture these interactions accurately, though, researchers also need to advance their simulations with very small time steps, meaning that they must recalculate particles’ positions and interactions at micro or nanosecond intervals. In order to do this, researchers need access to powerful enough processors to quickly solve these equations, but also a computer that has large amounts of memory that cores can continually access as well as fast connections between the individual computer cores so they may share their results with computer cores calculating the spaces nearby.
Modelling air moving in and out of the human lungs is, in principle, the same approach to modelling other fluid flows. Unfortunately, unlike modelling fluid flows in a uniform, stationary object like a fuel injector, lungs change shape while breathing, when air, the liquid that lines the inner surface of the lung, and tissue interact, and disperse inhaled air throughout increasingly smaller tubes before arriving at the alveoli where they can process the inhaled air into its constituent gases for use in the body.
To address these additional complications, the TUM team started to create novel models and develop more novel computational techniques. One of Wall’s collaborators, Dr. Martin Kronbichler, has been focused on high performance in CFD modelling for more than a decade. He recognized that while processors have gotten more powerful every few years, the memory bandwidth for rapidly sharing information between processors has not kept pace.
“Nowadays, modern machines are so powerful at computing, but there is this bottleneck with transferring data from memory to the computer cores,” he said. “So, we were observing that the movement is the expensive part, and we decided to try and use a so-called matrix-free algorithm, meaning that rather than saving information that we need for the next step then having to access it again, we’re just re-computing this information. While it required us to innovate and go away from how CFD software has traditionally been written, our new code ExaDG factors quicker because we can really utilize this method efficiently. So ultimately, we are computing more, but it is faster because you need to access less data.”
The time steps themselves also present a computational hurdle, as the more frequently a code has to recalculate the area in question, the less total time can be captured in a simulation. The team has been working on another indirect method to create more efficient calculations by optimizing each time step, allowing for more expensive individual time steps move forward in time in wider intervals. To date, the team was able to optimize its simulations to require less than 0.1 second per time step, and is focused on continuing to optimize the myriad memory bandwidth and latency issues to improve performance further.
Collaborations and combinations set the stage for better patient outcomes
Despite significant performance gains and a stable of relatively novel computational models and methods that could be explored for further improvements, Wall and his collaborators recognize hospitals are not on the cusp of buying supercomputers for themselves, and even the best-performing simulations are not fast enough to support individual recommendations quick enough to help seriously ill patients.
Ultimately, the need to find balance between accuracy and efficiency is endemic across most areas of research in computational (bio) mechanics. Researchers who have access to world-class computational resources such as those at LRZ tend to run computationally expensive high-resolution simulations that calculate as many parameters as possible “from scratch.” These simulations can then provide inputs for less computationally demanding simulations, making them more accurate and useful.
In the case of patients hospitalized with respiratory illnesses, every minute counts, and the team wants to use its computational work as the basis from which doctors could make more informed decisions about respirating a patient. Currently, doctors must use their background and training to predict what parameters to use for a ventilator, then modify and adjust on the fly based on whether the oxygen level in the blood is improving.
“In the coming years, we hope that our method can develop into providing a somewhat optimal ventilation strategy for each individual patient,” Wall said. “Doctors primarily base these parameters on the oxygen level in the blood, which is of course very important, but the lung is a very heterogeneous and complex organ, and while you might be getting the patient more oxygen, you could risk damaging the lungs through the ventilation process itself. And doctors currently have no way to see this at the moment. We want to have models available that, with a combination of CT-scan data and recording of patients’ breathing, can provide a suggestion for better ventilation parameters to use for that specific patient.” Wall and his collaborators have spun off this work into a start-up company, Ebenbuild GmbH.
Part of the success of this plan hinges on the team further optimizing its work on cutting-edge HPC resources. The team’s long-running collaboration with LRZ has not only provided the means to run simulations on a powerful supercomputer, but also resulted in regular exchange about the team’s computational needs more broadly. The team has worked closely with Dr. Momme Allalen, Leader of LRZ’s CFDLab team and user support specialist, as well as LRZ leadership. They have also had access to a variety of testbed systems at LRZ, most recently through its BEAST testbed.
“There are various levels of support we’ve received from LRZ,” Kronbichler said. “We have a lot of interaction with Momme to tune our algorithms for the architecture at hand. When you go to a new machine, you start to see new bottlenecks, and while they were likely already there on the machine before, you don’t notice them until you start leaving a lot of performance on the table. The collaboration with LRZ really helps us, because we have access to not just a single architecture, but multiple architectures.”
Additionally, the team collaborates closely with Prof. Dr. Martin Schulz, who in addition to his role as leading TUM’s Chair of Computer Architecture and Parallel Systems in the Informatics department, also works closely with LRZ and serves on the centre’s Board of Directors. The team works closely with Schulz to explain their hardware and software limitations and requirements, and tries to provide details that can inform future computational investments from the centre.
While the team recognizes that running optimal and fully comprehensive individualized lung simulations is not yet around the corner, it knows that optimizing today’s capabilities while planning improvements based on the promises of tomorrow’s computing resources will ultimately help researchers and patients alike.
-Eric Gedenk
Funding for SuperMUC-NG was provided by the Bavarian State Ministry of Science and the Arts and the German Federal Ministry of Education and Research through the Gauss Centre for Supercomputing (GCS).