LIFE SCIENCES
Molecular Simulations of Enzymes Under Reaction Conditions
Principal Investigator:
Jürgen Pleiss
Affiliation:
Institute of Biochemistry and Technical Biochemistry, University of Stuttgart (Germany)
Local Project ID:
Biocat
HPC Platform used:
Hazel Hen of HLRS
Date published:
Protein-substrate interactions are crucial for enzymatic activity. In the substrate binding site of an enzyme, the substrate is expected to bind in a productive orientation in close proximity to the active site residues of the enzyme. Scientists of the Institute of Biochemistry and Technical Biochemistry of the University of Stuttgart developed a simulation method to model substrate access to the active site of esterases, lipases, and acyl transferases. There is increasing experimental evidence that substrate access to and product exit from the binding site might become rate limiting steps under realistic reaction conditions. To study the concentration dependency of substrate access at high substrate concentrations. A series of simulations were performed for the enzymes with different substrates at varying concentrations, and the free energy profile was determined for substrate binding as a function of the distance of a substrate molecule from the active site of the enzyme.
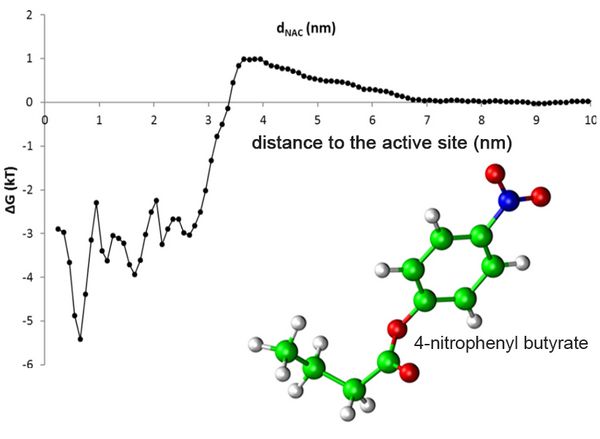
A typical substrate of Candida antarctica lipase B is 4-nitrophenyl butyrate (Fig. 1). The free energy profile demonstrated that at distances beyond 7 nm, the substrate does not interact with the enzyme, but binds to the protein surface and the substrate binding pocket, indicated by a drop of the free energy by 3.5 kT below a distance of 3.3 nm. Free energy profiles are a valuable tool for a better understanding of substrate access, to identify hotspots for protein engineering, and to support process design.
The GROMACS 5 software with the OPLS-AA all-atom force field was used. More than 500 simulations for different enzymes, substrates, and concentrations were performed. In total, more than 50 μs of molecular dynamics simulations were conducted on systems consisting up of 100,000 - 1,000,000 atoms using Hazel Hen of the High Performance Computing Center (HLRS) in Stuttgart, Germany.
Research Team:
Valerio Ferrario, Henrique Carvalho, Jürgen Pleiss - Institute of Biochemistry and Technical Biochemistry, University of Stuttgart
Reference:
Ferrario V, Pleiss J, 2019. Molecular simulations of enzymes under non-natural conditions. Eur. Phys. J. Special Topics 227: 1631 -1638
Scientific Contact:
Prof. Dr. Jürgen Pleiss
Institute of Biochemistry and Technical Biochemistry
University of Stuttgart
Allmandring 31, D-70569 Stuttgart (Germany)
e-mail: Juergen.Pleiss [@] itb.uni-stuttgart.de
http://www.itb.uni-stuttgart.de/
HLRS project ID: Biocat
Date published: February 2020